Chemical Structure Reconstruction
The Challenge: To develop an algorithm to determine the minimum number of descriptors required to reverse engineer a chemical structure.
Our Approach
We identifed 17 core chemical structural mordred descriptors that would allow a researcher to effectively deduce the structure of a molecule. Without any of these core structural descriptors, a researcher would not be able to accurately sythesize the structure. The remaining descriptors were identified as non-core morded descriptors.
We used a multiple output linear regression model. The model trains using all of the non-core descriptors to predict the value of the core descriptions. The model is versatile and is able to integrate other core structural descriptors as predictors, should the user choose to do so.
Methods
- We converted the data from .sdf files to. csv using the code provided with the data (sample data file and sdf to csv conversion provided in the sdf_to_csv file)
- We cleaned the data. We removed any irregularities from it, and extracted only nummberical values.
- We extracted the 17 core structural descriptors.
- We trained a Multiple Output Regressor to predict the 17 core structural descriptors. Using all non-core Mordred descriptors, the model was able to predict of all these descriptors with 100% 5-fold cross-validation accuracy.
- We iteratively thresholded the data, removing the descriptors with the least variance, to find the minimum number of descriptors for the model to accurately predict the 17 core structural descriptors.
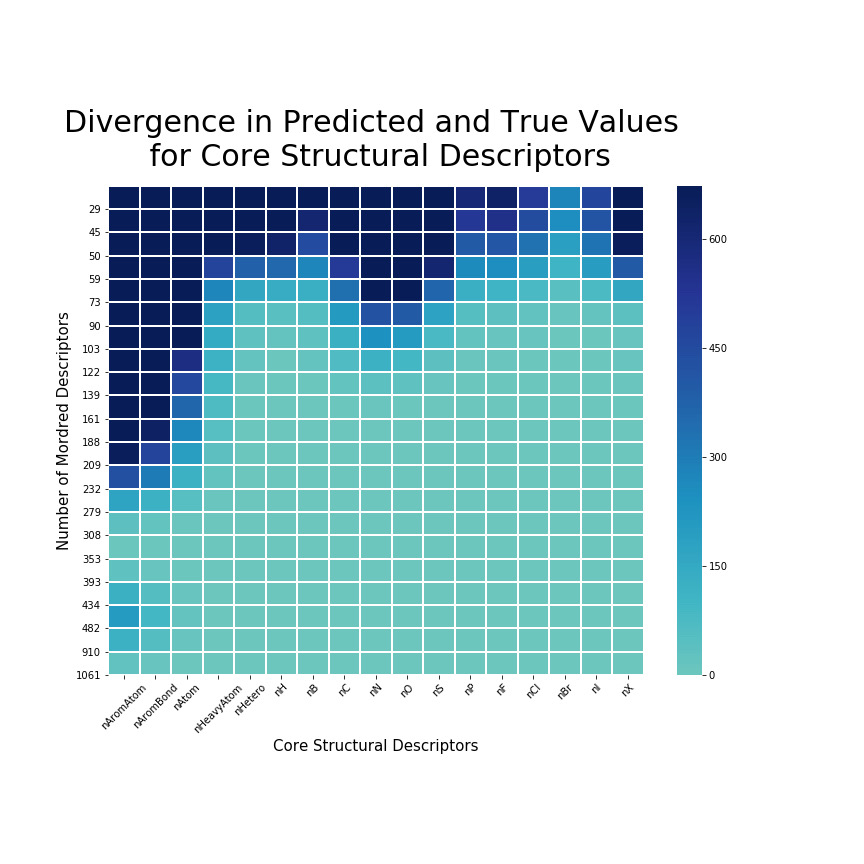
Results
Using less than around 50 Mordred desciptors, the model poorly predicts all 17 core mordred descriptors.
Try it out!
Go to Github Repo for this challenge
To run the model:
- Download the mordred compound sets (1 through 3) and place them in a data/ folder
- Open the src/config.py folder and change the data_path and results_path.
- Run the main function in src/thresholding.py.
- To visualize, run results_visualization.ipynb on jupyter notebook.
Notes
- Winning Project of Pharmahacks (2019) sponsored by Boehringer Ingelheim, Novartis, Student Pharma, Invivo AI, and MILA.
- This project included the exhaustive effort of five team members. Other descriptors were presented in the challenge but we chose to stick to mordred descriptors.

Comments